Fluorescence in
situ hybridization merupakan pendekatan metodologi yang terbaru dalam
pemeriksaan perubahan genetic pada sel tubuh.FISH mampu menditeksi amplifikasi
gen baik pada metaphase kromosom maupun pada interfase nuclei.
FISH dapat
menentukan level amplifikasi dan pola amplifikasi(clustered signals dan
multiple scattered signals) yang berhubungan sesuai dengan penguatan yang
terjadi pada homogeneous staining regions
dan double-minute chromosome.1,2,3
FISH dapat
menditeksi amplifikasi gen tidak hanya pada isolated
nuclei dan imprinted cells akan
tetapi dapat juga pada jaringan yang difiksasi formalin dan diblok paraffin
serta merupakan perangkat yang mantap untuk menditeksi amplifikasi gen pada
specimen tumor padat.2
FISH dapat
digunakan untuk menditeksi mikrodelesi,translokasi komplek dan perubahan
telomere yang tidak dapat diditeksi oleh karyotyping
rutin.1,2,3
FISH dapat juga
diaplikasikan untuk mengidentifikasi
pasien-pasien kanker payudara yang terpilih untuk diterapi dengan humanized monoclonal antibody terhadap
protein c-erbB2 (trastuzumab).2
Oleh karena
banyaknya aplikasi dari FISH,maka ada baiknya kita mengetahui secara mendasar
tentang prinsip kerja dari FISH.
Tinjauan Pustaka
Pengertian
FISH adalah
suatu teknik pemeriksaan sitogenetik yang merupakan suatu tipe hibridisasi yang
menggunakan complementary DNA atau RNA
strand (probe) yang dilabel
dengan bahan fluorescence untuk melokalisir rantai DNA|atau RNA yang spesifik
yang penampilannya dapat dilihat di bawah mikroskop fluorescence.1,2,3,4
Prinsip Kerja
FISH menggunakan
fluorescence probes yang akan
mengikat bagian dari kromosom (rantai DNA/RNA yang spesifik) yang menunjukkan
derajat rantai yang mirip.Ikatan ini kemudian dilihat di bawah mikroskop
fluorescence.1,2,3,4
Teknik Pemeriksaan
Pada pemeriksaan
FISH,kita menggunakan probes untuk menditeksi suatu gen yang spesifik yang
terdapat di nuclei yang terisolasi.Probes yang digunakan minimal 10 Kb atau
lebih dari itu.Probes yang kita jumpai di pasaran memiliki ukuran berkisar antara 30-100 Kb.Probes tersebut
terdiri dari 300 bases rantai DNA yang unik.Probes yang lebih pendek
menyebabkan proses hibridisasi yang tidak spesifik.
Beberapa probes
tersebut dilabel dengan biotin atau digoxigenin(indirectly labeled probes) dan
haptens ini kemudian diditeksi dengan streptoavidin atau anti-digoxigenin
antibody yang dilabel dengan bahan fluroscence seperti FITC (Fluoroscence
Isothiocyanate) atau rhodamine.FISH probes dari Vysis secara langsung dilabel
dengan bahan fluorescence seperti Spectrum Orange, Spectrum Green,
SpectrumAqua, dll. FISH probes tersebut dapat diproduksi dengan PCR jika kita
menginginkan rantai DNA yang lebih panjang dari 10 Kb.FISH probes yang
dihasilkan bersifat spesifik,seperti untuk K-sam terlokalisir pada 10q26
sebagai amplifikasi gen pada kanker lambung.2
FISH pada suspensi nuclear yang terisolasi
Jaringan tumor
yang telah diangkat sesegera mungkin dipotong sehalus-halusnya dan kemudian
diproses.Jaringan yang telah disiapkan ,diinkubasi selama 60 menit pada suhu 370C
di dalam Eagle’s minimal
essential medium yang mengandung 5% serum anak sapi dan 0.1%colagenase(protocol
1) ,selanjutnya di-vortex.Ataupun jaringan tersebut dapat diinkubasi dalam KCl
hipotonik(75 mmol/L) selama 15 menit pada suhu 370C(protocol2).
Setelah itu
difiltrasi melalui nylon mesh(50um) dan filtratnya dicuci sebanyak dua kali dengan phosphate-buffered
saline yang dicampur dengan Carnoy,s solution dan kemudian disimpan
pada suhu -200C sampai kita gunakan.
Bila
dibandingkan kedua protocol diatas,maka tidak terdapat perbedaan yang
berarti,walaupun pada protocol 2 diperlukan waktu yang lebih singkat.
Langkah
selanjutnya suspensi nuclear tadi dihapuskan pada silanized glass slide.2
FISH pada jaringan yang difiksasi formalin dan
diblok paraffin
Parameter
fiksasi seperti keterlambatan fiksasi,waktu fiksasi,pH ,konsentrasi
formalin,ukuran blok dan suhu pada saat proses blok paraffin sangat
mempengaruhi hasil FISH.
Tingkat
keberhasilan dalam pemeriksaan FISH tergantung pada pemisahan protein untuk
memperoleh DNA untuk hibridisasi.Metode digesti yang sering digunakan adalah
dengan sodium bisulfate/proteinase K.
Sayatan jaringan
yang telah diparaffinisasi dan direhidrasi diletakkan di silanized slides dan kemudian diinkubasi dalam 20% sodium
bisulfate/2xstandard saline citrate(2xSSC) pada suhu 430C selama 20
menit.Kemudian setelah dicuci dengan 2xSSC,pada slide diberikan proteinase
K(25ng/ml) dan diinkubasi pada suhu 370C selama 30 menit.Selanjutnya
dicuci lagi dengan 2xSSC dan didehidrasi dengan etanol,dikeringkan dan
didenaturasi dengan probe.2
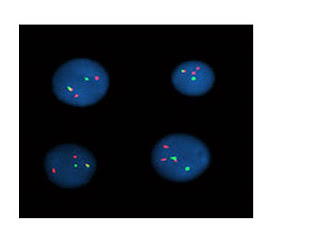
Amplifikasi onkogen pada sel-sel tumor
Pada sel-sel
mammalia,amplifikasi DNA yang tinggi dapat dijumpai pada dua struktur yaitu homogenously staining region (HSRs) dan double minute chromosome(DMs).HSRs
terlokalisasi di dalam suatu kromosom dalam bentuk meluas sepanjang chromosome region,sedangkan DMs
merupakan struktur bebas yang mengelilingi sentromer.
Amplifikasi gen
pada HSRs berupa clustered signals dan pada DMs berupa multiple scattered signals.
Pada pemeriksaan
FISH dapat digunakan single colour ataupun dual-colour.Pada single colour FISH
,dikatakan amplifikasi gen bila terlihat lebih dari 4 signal per nucleus.
Sedangkan pada
dual-colour FISH,digunakan gene specific
probe dan centromere specific probe
yang dilabel secara berbeda.Kemudian dihitung ratio onkogen signal terhadap
sentromer signal.Apabila rationya lebih besar dari 3 atau 4,maka dapat
dikatakan amplifikasi pada beberapa studi.2
1.Kumar,Abbas,Fausto.Pathologic
Basis of Disease.Seventh edition.Elsevier
Saunders.Philadelphia.2005
2.Oncogene Amplification Detection
by Fluorescence In Situ Hybridization,
available at :http://npg.nature.com/modpathol/keyword_index/kiiy.html
3.Fluorescence In Situ
Hybridization,available at:http://en.wikipedia.org/wiki/
Fluorescence_in_situ_hybridization
4.In Situ Hybridization,available
at:http://en.wikipedia.org/wiki/In_situ_hybridization
5.Fluorescence In Situ
Hybridization,available at:http://members.aol.com/chrom info
/fishinfo.htm
6.Protocol for Fluorescence In Situ
Hybridization,available at:http://www.hku.hk/
Oncology/lcg/ProtocolforFISH.htm
LAMPIRAN6
|
Protocol
for Fluorescence in situ Hybridization (FISH)
|
FISH is a very widely used technique on not only cytogenetic studies, but also other biological fields. It include metaphase & interphase FISH. This protocol will be divided into three parts: Probe labeling, Hybridization, and Washing.
v v
Probe labeling:
Several methods are used to do probe
labeling: Nick translation, Random Priming, and PCR.
Probes Labeling for FISH by Nick Translation
Materials:
Nick Translation Kit (Gibco, Cat#: 18247-015)
Components:
10X dNTP Mix
0.2 mM each dCTP, dGTP, dTTP
0.1 mM dATP
0.1 mM biotin-14-dATP
500 mM Tris-HCl (pH 7.8)
50 mM MgCl2
100 mM beta-mercaptoethanol
100 ug/ml nuclease-free BSA
10X Enzyme Mix
0.5 U/μl DNA Polymerase I
0.007 U/ul DNase I
50 mM Tris-HCl (pH 7.5)
5 mM magnesium chloride
0.1 mM phenylmethylsulfonyl fluoride
50% (v/v) glycerol
100 ug/ml nuclease-free BSA
Stop Buffer 0.5 M EDTA
(pH 8.0)
Distilled H2O
Procedures:
1. 1. Place a 0.6mL microcentrifuge tube on ice and allow the
tube to cool.
2. 2. Pipet the following components to the tube:
|
Volume
|
Reagent
|
|
5
mL
|
10X dNTP mix (minus
dATP)
|
|
5
mL
|
10X dATP +
Biotin-dATP
|
|
x
mL
|
1mg DNA (YAC, BAC)
|
|
35–
x mL
|
dH2O
|
|
45
mL
|
Total
volume
|
3. 3. Mix the tube briefly. Add 5uL Pol 10X Enzyme Mix. Mix thoroughly but gently. Centrifuge briefly in a centrifuge to bring
liquid to the bottom of the tube.
4. 4. Incubate at 15°C for 1-2 hours in a
PCR machine.
5. 5. At 1 hour, stop the reaction by placing the tubes in -20°C.
6. 6. Check the size of the labeled probes (2mL) by gel
electrophoresis in 0.7~2% agarose gel, looking for the peak size between 50 –
500 bp (or 100-300 bp) DNA fragments.
7. 7. If the size range is larger than this, add a further 5mL
enzyme mix, place at 15ºC for a further 30-60 min, and run another on a gel to
test the size.
8. 8. Stop the reaction by either adding the Stop Buffer or
heating in a 75°C water bath for 10
minutes (or incubate at 75°C in a PCR machine).
9. 9. Chill on ice.
10.
10. Combine
the following in a 1.5mL microcentrifuge tube:
48mL Biotin-labeled DNA
1mL glycogen or 50mg salmon sperm DNA
11.
11. Then,
add 51mL
dH2O to make up to 100mL.
12.
12. Add
0.1 volume 3M Sodium Acetate solution (pH 5.6) into the DNA sample; e.g., 100mL DNA solution + 10mL 3M Sodium Acetate
solution.
13.
13. Add
2.5X volume (250mL) cold absolute ethanol to the mixture, mix well.
14.
14. Incubate
the mixture at –20°C for at least 30-60
minutes.
15.
15. Centrifuge
the mixture for 20 minutes at highest speed at 4°C.
16.
16. Discard
the supernatant, then vacuum dry the DNA for about 10 minutes.
17.
17. Store
the DNA probe in dry form at –20°C until use.
18.
18. Add
x mL
of dH2O (usually 10-20 mL) to the precipitated DNA (according
to the size of the pellet). To give a final concentration of 50ng/mL. Allow the DNA to dissolve at RT for 1-2 h or
at 4°C overnight with
occasional mixing. Purified, labeled
probes are stable for several years stored at –20°C.
Probes Labeling for FISH by Random Priming
Method
Materials:
BioPrime DNA Labeling System (Cat#:
18094-011)
Components:
2.5X Random Primers
Solution:
[125 mM Tris-HCl (pH
6.8), 12.5 mM MgCl2, 25 mM
2-mercaptoethanol, 750 ug/ml oligodeoxyribonucleotide primers (random octamers)]
10X dNTP Mixture:
[1 mM biotin-14-dCTP, 1 mM dCTP, 2 mM dATP, 2
mM dGTP, 2 mM dTTP in 10 mM Tris-HCl (pH 7.5), 1 mM Na2EDTA]
Klenow Fragment (Large
Fragment of DNA Polymerase I):
[40 U/ul Klenow Fragment in
50 mM Potassium Phosphate (pH 7.0), 100 mM KCl,
1 mM DTT, 50%
Glycerol]
Stop Buffer: [0.5 M Na2EDTA (pH 8.0)]
Distilled Water
Procedures:
1.
Dissolve 100 ng DNA in 5-20 ul of dilute buffer in a microcentrifuge tube. On
ice, add 20 μl 2.5X Random Primers Solution.
2. 2. Denature by heating for 5 min in a boiling water bath;
immediately cool on ice. (The amount of template per reaction has been varied
from 25-500 ng with satisfactory results.)
Perform the following additions on ice:
5 ul 10X dNTP Mixture
Distilled Water to a total volume of 49 ul
3. 3. Mix briefly.
4. 4. Add 1 ul Klenow Fragment. Mix gently but thoroughly.
Centrifuge 15-30 sec.
5. 5. Incubate at 37°C for 60 min.
6. 6. Add 5 ul Stop Buffer.
Probes Labeling for FISH by DOP-PCR: (see
Protocol of Chromosome Microdissection)
v v
Hybridization:
1.
Materials:
1.1. Slide
Pretreatment
1. (Optional: not recommended for
metaphase FISH): PK stock solution: 5 mg proteinase K (Boehringer,
Mannheim, Germany), 50 mL 1M Tris-HCl (pH 7.5), 20 mL 0.5M EDTA (pH 7.0),
2 mL
5M NaCl, make up to 1 mL in filtered
double- distilled water; make fresh as required.
2. 20X
standard saline citrate (SSC) stock solution: 3.0M NaCl, 0.3M Na-citrate;
set up with double-distilled water, adjust to pH 7.0, autoclave, and store at
room temperature.
3. RNase
stock solution: 10 mg/mL of RNase type A (Boehringer); set up with filtered
double- distilled water; aliquot and store at -20ºC.
4. RNase
solution: per slide 200 mL 2X SSC plus 10 mL of RNase stock solution are
necessary; make fresh as required.
5. (Optional:
not recommended for metaphase FISH): Pepsin stock solution 10% (w/v):
dissolve 100 mg pepsin (Serva, Heidelberg, Germany) in 1 mL of filtered double-distilled water at 37ºC; aliquot and store at
-20ºC.
6. (Optional: not recommended for metaphase
FISH): Pepsin buffer: Add 1 mL of
1M HCl to 99 mL of distilled water
and incubate at 37ºC for about 20 min; then add 50 mL of the pepsin stock
solution 10% (wlv) and leave the coplin jar at 37ºC; make fresh as required.
7.
7. (Optional:
not recommended for metaphase FISH): 1X PBS/ MgCl2: 5% (v/v) 1M MgCl2
in 1X PBS. (2.5mL 1M MgCl2 in 47.5mL 1X PBS)
8.
8. (Optional:
not recommended for metaphase FISH): Formalin buffer: 3% (v/v) of acid-free
formaldehyde (37%; Roth) in 1X PBS; make fresh as required.
1.2. Fluorescence In
Situ Hybridization (FISH)
1.2.1. Slide Denaturation
1.
1. Denaturation buffer(preferred): 70% (v/v)
deionized formamide, 10% (v/v) filtered double-distilled water, 10% (v/v) 20X
SSC, 10% (v/v) phosphate buffer; make fresh as required.
OR: Denature
solution: 70% (v/v) formamide, 2X SSC (pH7.0), 0.1mM EDTA, pH7.0
Add 175mL formamide, 25mL 20X SSC (pH7.0), 50mL 0.5M EDTA, pH7.0
and 50mL purified H2O to make 250mL solution and mix
thoroughly. Verify that the pH is
7.0-7.5 by measuring the pH at ambient temperature. Between use, store covered at 4°C.
Discard after 7 days.
2. Deionized formamide: Add 5 g of ion
exchanger Amberlite MB1 (Serva) to 100 mL of formamide (Merck, Darmstadt,
Mannheim, Germany) stir for 2 h (room temperature) and filter twice through
Whatmann no. 1 filter paper. Aliquot and store at -20ºC.
3.
Phosphate buffer: prepare 0.5M Na2HPO4 and 0.5M NaH2PO4,
mix these two solutions (1: 1) to get pH 7.0, then aliquot and store at -20ºC.
1.2.2. Probe Denaturation
1. Hybridization buffer: Dissolve 2 g dextran
sulfate in 10 mL 50% deionized formamide/2X SSC/50 mM phosphate buffer for 3 h at 70ºC. Aliquot and store at -20ºC.
OR: Hybridization
solution: MM2.1: 5.5mL formamide
1g Dextran sulfate
0.5mL 20X SSC
Heat to 70°C for several hours to dissolve the dextran
sulfate, then cool and adjust to pH 7.0 and add water to volume of 7mL.
1.2.3. Posthybridization and Detection Washing
1.
1. Washing solution I: 50% (v/v) formamide
(Merck), 10% (v/v) 20X SSC, 40% (v/v) distilled water; make fresh as required.
2.
2. Washing solution 2 (WS-2)(4X SSC/0.05% Tween
20).
3.
3. Washing solution 3 (WS-3)(4X SSC).
4.
4. Blocking solution: 1-3% (w/v) BSA in 4X SSC,
0.05% (v/v) Triton X-100 (make up fresh).
5.
5. PN buffer: 0.1M NaH2PO4/0.1M
Na2HPO4 M, pH 8.0; 0.1% NP-40.
6.
6. PNM buffer:
Add
5% (w/v) non fat dry milk to PN buffer plus 0.02% (w/v) Na-azide, incubate at
37°C overnight. It will look terrible. Centrifuge the solution for 5 minutes at
1000g. Transfer the supernatant to a clean tube and store at 4°C.
7.
7. FITC-Avidin (2mg/2mL): Add 398mL PNM
buffer to make up 5mg/mL.
OR: Solution
1: FITC-avidin (CAMON Vector Laboratories)/4X SSC/0.2 %Tween/5% BSA (1: 300
both Sigma, St. Louis , MO
8.
8. Anti-Avidin (2mg/2mL): Add 398mL PNM
buffer to make up 5mg/mL.
OR: Solution ll: Biotinylated antiavidin (CAMON
Vector Laboratories)/Anti-digoxigenin- rhodan-dne (Boehringer Mannheim , Germany
9.
9. Antifade solution
100mg p-phenylenediamine dihydrochloride in
10mL PBS. Adjust to pH 8.0 with 0.5M
carbonate-bicarbonate buffer (0.42g NaHCO3 in 10mL dH2O,
adjust pH to 9.0 with NaOH). Add to 90mL
with glycerol. Filter with 0.22m membrane to remove
undissolved particals. If necessary, add
0.5-1mg/mL
DAPI.
OR: DAPI-solution:
Dissolve 5 mL
of DAPI (4,6-diamidino-2-phenylindol.2HCl stock- solution; Serva) in 100 mL 4X
SSC/0.2% Tween; make fresh as required.
OR: 70%
(v/v) deionized formamide, 10% (v/v) filtered double-distilled water, 10% (v/v)
20X SSC, 10% (v/v) phosphate buffer, make fresh as required.
2.
Procedures for Fluorescence In Situ Hybridization (FISH):
Pre-treatment of slides
1. 1. (Optional) If the slide is not dry enough, dehydrate the
slide by immersing the slide into 100% ethanol for 1 min. Air dry.
2. 2. Pretreat slide with 200mL diluted RNase
solution (0.1mg/mL) for about 30 to 60 minutes at 37°C.
3. 3. Wash slide with 2X SSC for 5 minutes (with agitation).
4. 4. Pepsin treatment: (Optional, if the chromosome targets
are bone marrow smear, bone marrow progenitors from methyl cellulose-grown
colony assays, tumour preparations)
i.
i. Add 200mL diluted (with 0.01M
HCl) pepsin to the slide
ii. ii. Incubate slides at 37°C for 5-10 minutes.
iii. iii. Wash 2X with 1X PBS
for 5 min at RT with shaking.
Note:
over-digestion can also cause problems (loss of cells from the slide), so only
use when absolutely necessary.
5. 5. Place slides in PBS/50mM MgCl2 for 5 min.
6. 6. (Optional) Fix in PBS/50mM MgCl2/1%
formaldehyde for 10 min.
7. 7. Wash in PBS for 5 min (with agitation).
8. 8. Dehydrate the slide with 70%, 90%, and 100% ethanol for
1-2 min each and allow to air dry.
Slides can be stored desiccated at 4°C for up to one month
before use.
Pre-hybridization of DNA probes
9. 9. During incubation step 1, prepare hybridization mixture
as follows:
i.
i. Add 2mL (100ng) diluted
Biotin-labeled DNA + 1mL (2.5mg) Cot-1 DNA + 7mL MM2.1(warmed to
RT).
ii.
ii. Denature the hybridization mix at 75°C for 5-7 minutes.
iii. iii. Place the probe on ice for 3-5 mins. (very IMPORTANT!)
iv. iv. Transfer the denatured probe to 37°C for 15 min – 2 h for prehybridization.
Denature metaphase
chromosome slides
10.
10. Incubate slides in
denaturing solution (in water-bath) for 2
minutes at 75°C.
11.
11. Wash slides in cold 2X SSC, followed by two changes of
2X SSC.
12.
12. Dehydrate the slide
through a cold alcohol series (70%,
90%, and 100% ethanol for 1 min each)..
13.
13. Air dry the slides
and place on a hot plate at ~42°C.
14.
14. Apply 10mL of denatured probe
mix to the slide.
15.
15. Immediately apply a
coverslip and seal with rubber cement.
Keep the slide in a moist chamber at 37°C overnight – four
days for hybridization.
Washing slides
16.
16. Place the wash tanks
containing WS-1 in a 45°C water bath for at
least 30 minutes prior to use.
17.
17. Remove rubber cement
by using forceps. Coverslips can then be
removed either by soaking in 2X SSC or gently tipping them off into the glass
disposal bin (never pull them off!).
18.
18. Wash the slides 3
times with WS-1 at 45°C for 5 –10 min each
(usu. 5min).
19.
19. Wash the slides 3
times with WS-2 at RT for 2 min each.
20.
20. (Optional) a) Blocking treatment with 1-3% BSA in
4X SSC for 20 min at RT.
21.
21. b) Wash
22.
22. Wash the slides with
WS-3 for 2 min at RT.
Note: make sure the
slides are not dried in any point during the detection and washing steps.
Signal enhancement
23.
23. Add 40uL Avidin-FITC
(5ug/mL Avidin in PNM buffer) onto slides and cover with coverslip. Keep the slides in a moist chamber in dark
for 20 min at RT.
24.
24. Wash the slide as
steps 19, 21 (do not perform step 18 & 20). Then, proceed from 24 to 26
steps or directly jump to steps 27-29.
25.
25. (Optional): Add 40uL
anti-Avidin (5ug/mL anti-Avidin in PNM buffer) onto slides and cover with
coverslip. Keep the slides in a moist
chamber in dark for 20 min at RT.
26.
26. Wash the slides as
steps 23.
27.
27. Repeat steps 22-23 for
one additional Avidin-FITC treatment.
Visualizing the hybridization
28.
28.
Dehydrate the slides with 70%, 90%, and 100% ethanol for 1-2 min
each and allow to air dry.
29.
29.
Apply 40uL of DAPI II counterstain and a coverslip to
hybridization location.
30.
30.
Store in dark if not use, otherwise, examine the slide at once
under fluorescence microscope.




Mohon info untuk laboratorium yang pernah melakukan FISH ini di lab mana ya? Terimakasih
BalasHapusMohon info untuk laboratorium yang pernah melakukan FISH ini di lab mana ya? Terimakasih
BalasHapus